convert between bioinformatics formats
Project description
Bioconvert
Bioconvert is a collaborative project to facilitate the interconversion of life science data from one format to another.







- contributions:
Want to add a convertor ? Please join https://github.com/bioconvert/bioconvert/issues/1
- issues:
Overview
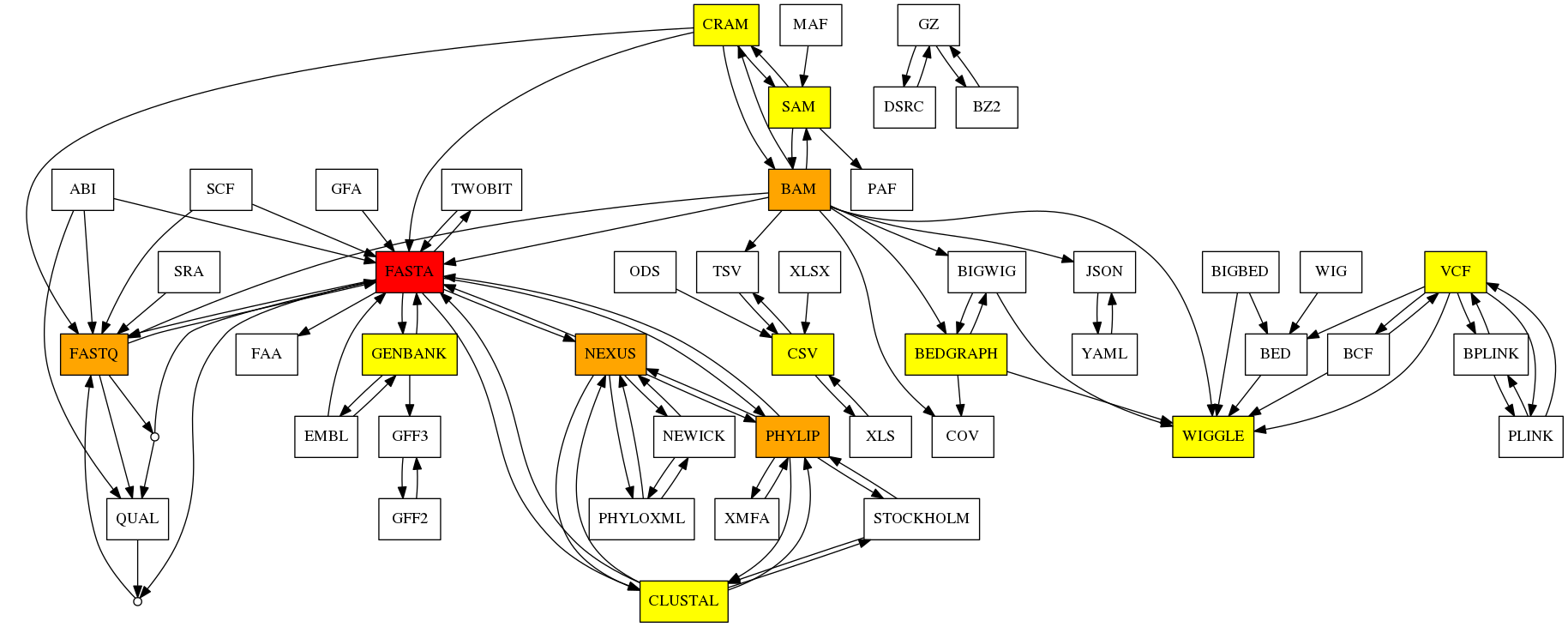
Life science uses many different formats. They may be old, or with complex syntax and converting those formats may be a challenge. Bioconvert aims at providing a common tool / interface to convert life science data formats from one to another.
Many conversion tools already exist but they may be dispersed, focused on few specific formats, difficult to install, or not optimised. With Bioconvert, we plan to cover a wide spectrum of format conversions; we will re-use existing tools when possible and provide facilities to compare different conversion tools or methods via benchmarking. New implementations are provided when considered better than existing ones.
In March 2022, we had 48 formats, 98 direct conversions (125 different methods).
Installation
In order to install bioconvert, you can use pip:
pip install bioconvert
We also provide releases on bioconda (http://bioconda.github.io/):
conda install bioconvert
and Singularity container are available. See http://bioconvert.readthedocs.io/en/master/user_guide.html#installation for details.
Usage
From the command line, you can convert a FastQ file into a FastA file as follows (compressed or not):
bioconvert fastq2fasta input.fastq output.fasta bioconvert fastq2fasta input.fq output.fasta bioconvert fastq2fasta input.fq.gz output.fasta.gz bioconvert fastq2fasta input.fq.gz output.fasta.bz2
When there is no ambiguity, you can be implicit:
bioconvert input.fastq output.fasta
For help, just type:
bioconvert --help bioconvert fastq2fasta --help
From a Python shell:
# import a converter from bioconvert.fastq2fasta import FASTQ2FASTA # Instanciate with infile/outfile names convert = FASTQ2FASTA(infile, outfile) # the conversion itself convert()
Available Converters
Converters |
CI testing |
Benchmarking |
|---|---|---|

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
||

|
Contributors
Setting up and maintaining Bioconvert has been possible thanks to users and contributors. Thanks to all:

Changes
Version |
Description |
|---|---|
0.5.1 |
|
0.5.0 |
|
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.











